ppdiag is an R package which provides a collection of tools which can be used to assess the fit of temporal point processes to data.
These currently include:
- Simulating data from a specified point process
- Fitting a specified point process model to data
- Evaluating the fit of a point process model to data using several diagnostic tools
Installation
You can install the released version of ppdiag from CRAN with:
install.packages("ppdiag")The current development version of this package is available from GitHub with:
# install.packages("remotes")
remotes::install_github("OwenWard/ppdiag")Example
To illustrate some of the basic functionality of this package, we can simulate data from a specified Hawkes process and examine our diagnostic results when we fit a homogeneous Poisson process to this data.
library(ppdiag)
hp_obj <- pp_hp(lambda0 = 0.2, alpha = 0.35, beta = 0.8)
sim_hp <- pp_simulate(hp_obj, end = 200)
sim_hp
#> [1] 5.760803 8.948532 9.985766 11.144890 11.305228 17.509538
#> [7] 21.990103 25.754804 25.756725 26.663477 30.420693 34.465694
#> [13] 36.599366 41.410990 44.438180 44.712109 45.211409 46.512076
#> [19] 49.511806 51.046608 52.335976 75.660840 76.639780 85.685473
#> [25] 87.284500 87.448896 87.722795 87.856140 89.034916 89.056869
#> [31] 89.179466 89.195399 90.775515 90.792069 91.042053 92.293751
#> [37] 92.355206 96.928276 97.053098 97.880772 98.310011 98.881077
#> [43] 99.010649 99.213401 99.347277 100.022472 100.679788 100.694527
#> [49] 101.452972 103.926526 106.653150 113.212708 113.738940 114.338943
#> [55] 114.745921 115.058328 115.128879 115.896050 116.130131 116.241804
#> [61] 118.427450 123.380339 123.502200 123.973237 128.881506 129.263522
#> [67] 143.028509 151.006471 158.349391 162.063008 168.034982 170.657158
#> [73] 171.781831 175.347477 175.762528 176.398255 176.780393 182.373018
#> [79] 191.028275 191.257033 192.095983 196.416187 197.413321We can readily evaluate the fit of a homogeneous Poisson process to this data.
est_hpp <- fithpp(sim_hp)
est_hpp
#> $lambda
#> [1] 0.4204377
#>
#> $events
#> [1] 5.760803 8.948532 9.985766 11.144890 11.305228 17.509538
#> [7] 21.990103 25.754804 25.756725 26.663477 30.420693 34.465694
#> [13] 36.599366 41.410990 44.438180 44.712109 45.211409 46.512076
#> [19] 49.511806 51.046608 52.335976 75.660840 76.639780 85.685473
#> [25] 87.284500 87.448896 87.722795 87.856140 89.034916 89.056869
#> [31] 89.179466 89.195399 90.775515 90.792069 91.042053 92.293751
#> [37] 92.355206 96.928276 97.053098 97.880772 98.310011 98.881077
#> [43] 99.010649 99.213401 99.347277 100.022472 100.679788 100.694527
#> [49] 101.452972 103.926526 106.653150 113.212708 113.738940 114.338943
#> [55] 114.745921 115.058328 115.128879 115.896050 116.130131 116.241804
#> [61] 118.427450 123.380339 123.502200 123.973237 128.881506 129.263522
#> [67] 143.028509 151.006471 158.349391 162.063008 168.034982 170.657158
#> [73] 171.781831 175.347477 175.762528 176.398255 176.780393 182.373018
#> [79] 191.028275 191.257033 192.095983 196.416187 197.413321
#>
#> attr(,"class")
#> [1] "hpp"
pp_diag(est_hpp, events = sim_hp)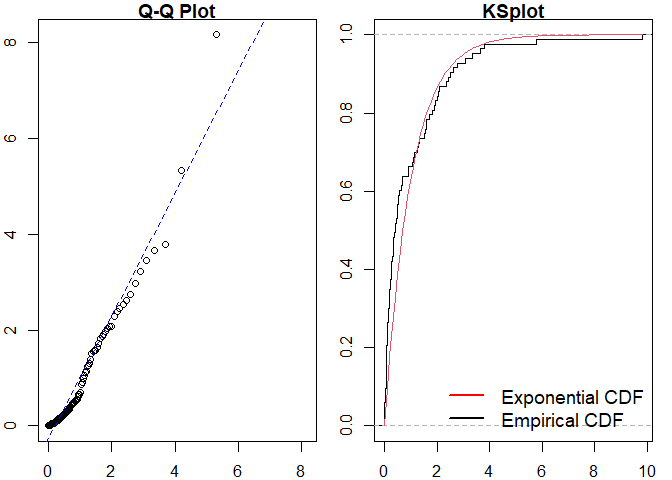
#>
#> Raw residual: 0
#> Pearson residual: -2.842171e-14
#>
#> One-sample Kolmogorov-Smirnov test
#>
#> data: r
#> D = 0.18659, p-value = 0.00529
#> alternative hypothesis: two-sided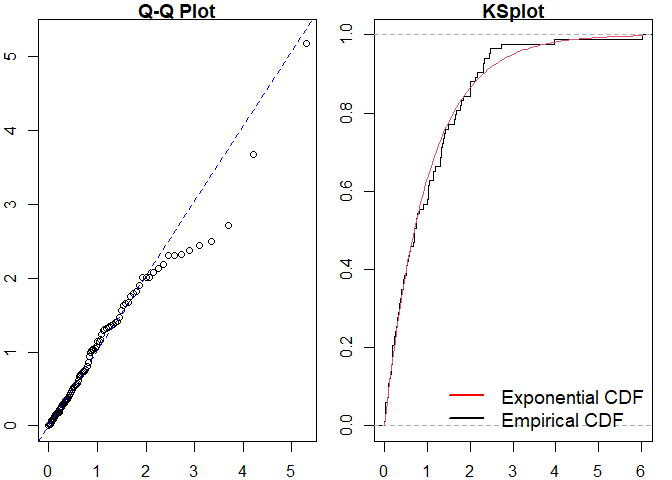
#> Raw residual: -0.005375166
#> Pearson residual: -0.3651356
#>
#> One-sample Kolmogorov-Smirnov test
#>
#> data: r
#> D = 0.064572, p-value = 0.8571
#> alternative hypothesis: two-sidedMarkov Modulated Hawkes Process Example
This is particularly useful for more complex point processes, such as the Markov Modulated Hawkes Process (MMHP). We can simulate events from this model and examine the fit of simpler point processes to this data.
Q <- matrix(c(-0.2, 0.2, 0.1, -0.1), ncol = 2, byrow = TRUE)
mmhp_obj <- pp_mmhp(Q, delta = c(1 / 3, 2 / 3),
lambda0 = 0.2,
lambda1 = .75,
alpha = 0.4,
beta = 0.8)
mmhp_obj
#> $Q
#> [,1] [,2]
#> [1,] -0.2 0.2
#> [2,] 0.1 -0.1
#>
#> $delta
#> [1] 0.3333333 0.6666667
#>
#> $events
#> NULL
#>
#> $lambda0
#> [1] 0.2
#>
#> $lambda1
#> [1] 0.75
#>
#> $alpha
#> [1] 0.4
#>
#> $beta
#> [1] 0.8
#>
#> attr(,"class")
#> [1] "mmhp"
mmhp_events <- pp_simulate(mmhp_obj, n = 50)We can easily fit a homogeneous Poisson process and visualise the goodness of fit.
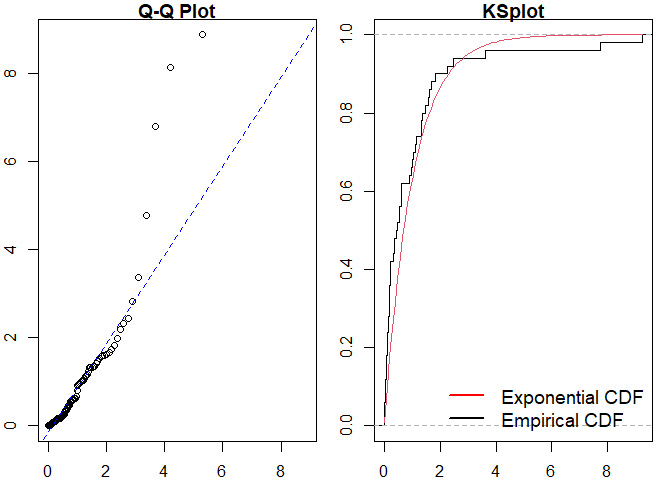
#>
#> Raw residual: -1
#> Pearson residual: -0.9740881
#>
#> One-sample Kolmogorov-Smirnov test
#>
#> data: r
#> D = 0.21168, p-value = 0.01914
#> alternative hypothesis: two-sidedSimilarly for a Hawkes process.
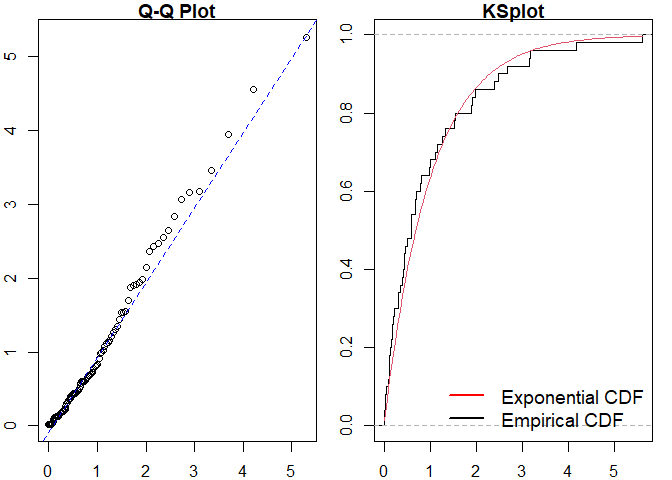
#> Raw residual: -0.3466309
#> Pearson residual: -0.6079909
#>
#> One-sample Kolmogorov-Smirnov test
#>
#> data: r
#> D = 0.095638, p-value = 0.7142
#> alternative hypothesis: two-sidedWe can then compare to the true point process model.
pp_diag(mmhp_obj, mmhp_events$events)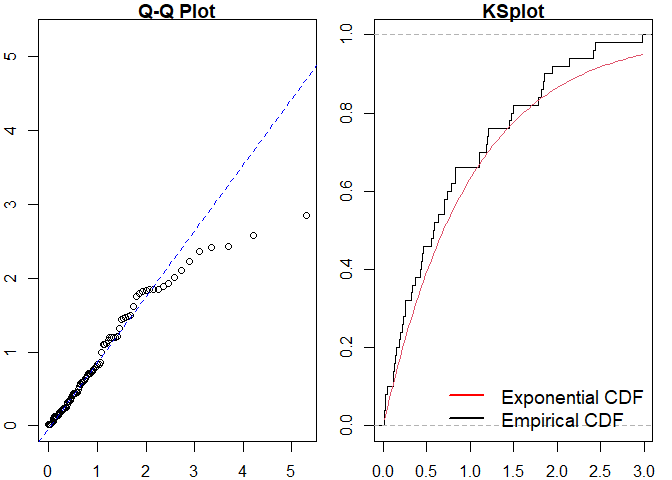
#> Raw residual: 8.862914
#> Pearson residual: 10.62449
#>
#> One-sample Kolmogorov-Smirnov test
#>
#> data: r
#> D = 0.096698, p-value = 0.7017
#> alternative hypothesis: two-sidedGetting help and contributing
Please file any issues here. Similarly, we would be delighted if anyone would like to contribute to this package (such as adding other point processes, kernel functions). Feel free to take a look here and reach out with any questions.
References
- Sun et al., (2021). ppdiag: Diagnostic Tools for Temporal Point Processes. Journal of Open Source Software, 6(61), 3133, https://doi.org/10.21105/joss.03133
- Wu et al., Diagnostics and Visualization of Point Process Models for Event Times on a Social Network, https://arxiv.org/abs/2001.09359
